Research
Biomaterials
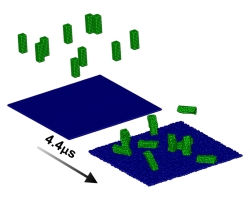
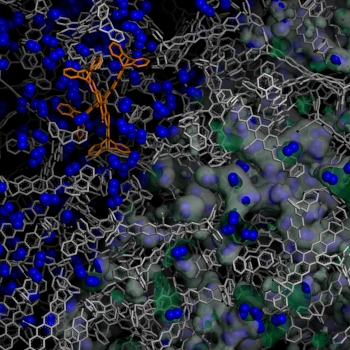
We are interested in understanding the relationship between movement and function in a protein, enzyme, virus, or aggregates of them. The tendency for globular proteins to aggregate causes their viscosity to increase with concentration much more rapidly than expected by simple models. Additionally, protein adsorption behavior has been a topic of intense research for decades, but a fundamental understanding remains uncertain. We utilize atomisic and mesoscale molecular simulations to investigate these phenomena.


Ionic Liquids
Ionic liquids are being studied for oil recovery and extraction of oil from tar sands. Deep eutectic solvents, which are ionic liquid analogues, show promise as novel solvents for these applications because they are cheaper than typical ionic liquids and are environmentally benign. We are utilizing molecular simulations to futher understand the thermodynamic and transport properties in these systems.
Nanoporous Materials
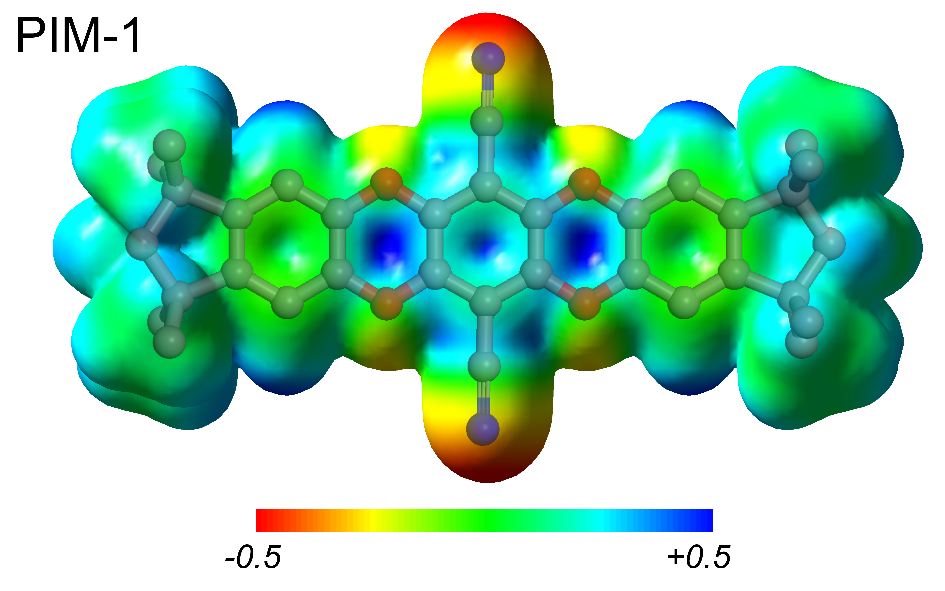
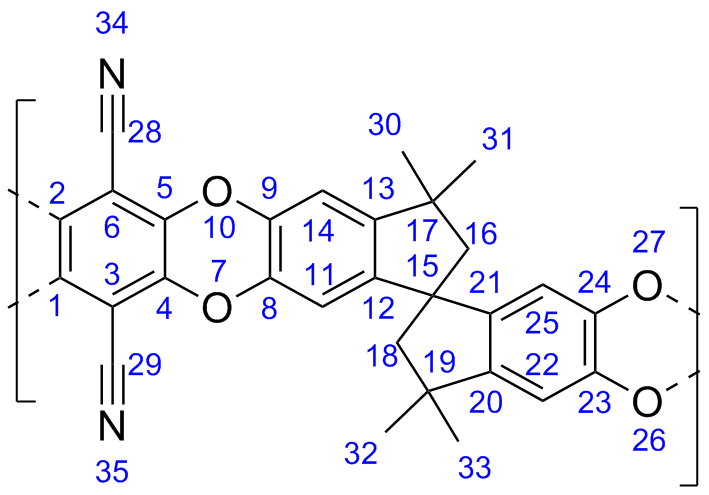
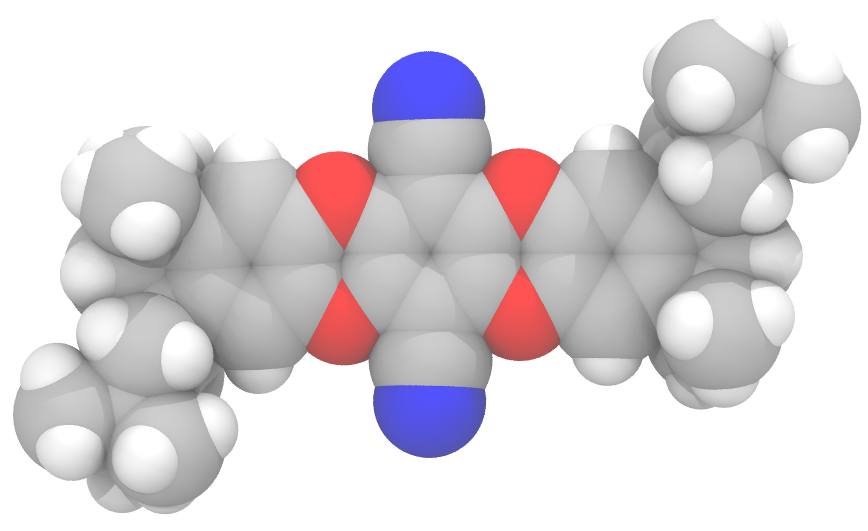
Organic microporous materials offer a new approach for environmentally conscious and energy efficient gas storage and separation technology. Our research focuses on polymers of intrinsic microporosity, organic molecules of intrinsic microporosity, and hypercrosslinked polymers. Utilizing molecular simulations, we can provide atomistic understanding of the structure and porosity of materials to help interpret experimental data and aid in the design of novel nanoporous polymers.

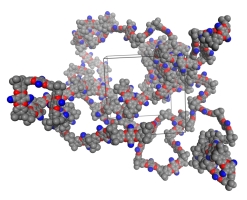

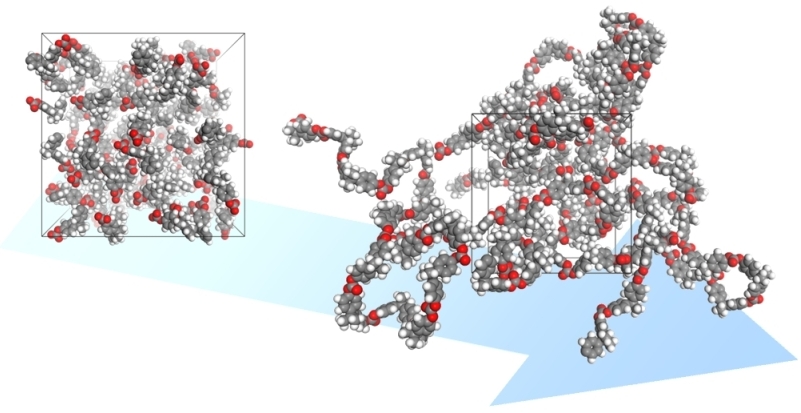
Polymatic
Amorphous materials present challenges to consistent and predictive structure generation in molecular simulations due to their complex and unordered nature. One focus of our research has been on the development of a general methodology for virtual synthesis of amorphous materials knowing only their chemical structures, called Polymatic. It is composed of two parts: (i) a simulated polymerization algorithm and (ii) a 21-step molecular dynamics compression and relaxation scheme.
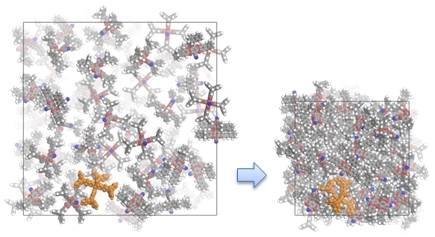
Force Field Database
This database is a collection of the molecular models used in some publications of the Computational Biophysics and Soft Materials group at Penn State. A molecule may be selected to obtain information on the force field functional forms and parameters used in published articles. For each model, you may download data files containing all necessary molecular modeling information.